Dimensionality reduction
Timothy Keyes
2024-08-25
Source:vignettes/dimensionality-reduction.Rmd
dimensionality-reduction.RmdA useful tool for visualizing the phenotypic relationships between single cells and clusters of cells is dimensionality reduction, a form of unsupervised machine learning used to represent high-dimensional datasets in a smaller number of dimensions.
tidytof includes several dimensionality reduction
algorithms commonly used by biologists: Principal component analysis
(PCA), t-distributed stochastic neighbor embedding (tSNE), and uniform
manifold approximation and projection (UMAP). To apply these to a
dataset, use tof_reduce_dimensions().
Dimensionality reduction with
tof_reduce_dimensions().
Here is an example call to tof_reduce_dimensions() in
which we use tSNE to visualize data in tidytof’s built-in
phenograph_data dataset.
data(phenograph_data)
# perform the dimensionality reduction
phenograph_tsne <-
phenograph_data |>
tof_preprocess() |>
tof_reduce_dimensions(method = "tsne")
#> Loading required namespace: Rtsne
# select only the tsne embedding columns
phenograph_tsne |>
select(contains("tsne")) |>
head()
#> # A tibble: 6 × 2
#> .tsne1 .tsne2
#> <dbl> <dbl>
#> 1 1.74 17.0
#> 2 10.4 7.60
#> 3 30.4 19.8
#> 4 15.2 14.6
#> 5 3.99 19.0
#> 6 21.3 12.4By default, tof_reduce_dimensions will add
reduced-dimension feature embeddings to the input tof_tbl
and return the augmented tof_tbl (that is, a
tof_tbl with new columns for each embedding dimension) as
its result. To return only the features embeddings themselves, set
augment to FALSE (as in
tof_cluster).
phenograph_data |>
tof_preprocess() |>
tof_reduce_dimensions(method = "tsne", augment = FALSE)
#> # A tibble: 3,000 × 2
#> .tsne1 .tsne2
#> <dbl> <dbl>
#> 1 18.7 -1.60
#> 2 9.55 3.71
#> 3 11.3 27.9
#> 4 14.8 13.4
#> 5 20.9 0.568
#> 6 23.3 14.5
#> 7 19.0 6.40
#> 8 27.1 14.7
#> 9 20.1 12.6
#> 10 12.7 -1.41
#> # ℹ 2,990 more rowsChanging the method argument results in different
low-dimensional embeddings:
phenograph_data |>
tof_reduce_dimensions(method = "umap", augment = FALSE)
#> # A tibble: 3,000 × 2
#> .umap1 .umap2
#> <dbl> <dbl>
#> 1 -5.17 -5.17
#> 2 -5.76 -4.26
#> 3 -7.72 0.819
#> 4 -6.54 0.271
#> 5 -4.95 -4.90
#> 6 -0.380 3.52
#> 7 -4.76 -4.44
#> 8 -7.57 1.40
#> 9 -6.43 -0.919
#> 10 -6.92 -6.41
#> # ℹ 2,990 more rows
phenograph_data |>
tof_reduce_dimensions(method = "pca", augment = FALSE)
#> # A tibble: 3,000 × 5
#> .pc1 .pc2 .pc3 .pc4 .pc5
#> <dbl> <dbl> <dbl> <dbl> <dbl>
#> 1 -2.77 1.23 -0.868 0.978 3.49
#> 2 -0.969 -1.02 -0.787 1.22 0.329
#> 3 -2.36 2.54 -1.95 -0.882 -1.30
#> 4 -3.68 -0.00565 0.962 0.410 0.788
#> 5 -4.03 2.07 -0.829 1.59 5.39
#> 6 -2.59 -0.108 1.32 -1.41 -1.24
#> 7 -1.55 -0.651 -0.233 1.08 0.129
#> 8 -1.18 -0.446 0.134 -0.771 -0.932
#> 9 -2.00 -0.485 0.593 -0.0416 -0.658
#> 10 -0.0356 -0.924 -0.692 1.45 0.270
#> # ℹ 2,990 more rowsMethod specifications for tof_reduce_*() functions
tof_reduce_dimensions() provides a high-level API for
three lower-level functions: tof_reduce_pca(),
tof_reduce_umap(), and tof_reduce_tsne(). The
help files for each of these functions provide details about the
algorithm-specific method specifications associated with each of these
dimensionality reduction approaches. For example,
tof_reduce_pca takes the num_comp argument to
determine how many principal components should be returned:
# 2 principal components
phenograph_data |>
tof_reduce_pca(num_comp = 2)
#> # A tibble: 3,000 × 2
#> .pc1 .pc2
#> <dbl> <dbl>
#> 1 -2.77 1.23
#> 2 -0.969 -1.02
#> 3 -2.36 2.54
#> 4 -3.68 -0.00565
#> 5 -4.03 2.07
#> 6 -2.59 -0.108
#> 7 -1.55 -0.651
#> 8 -1.18 -0.446
#> 9 -2.00 -0.485
#> 10 -0.0356 -0.924
#> # ℹ 2,990 more rows
# 3 principal components
phenograph_data |>
tof_reduce_pca(num_comp = 3)
#> # A tibble: 3,000 × 3
#> .pc1 .pc2 .pc3
#> <dbl> <dbl> <dbl>
#> 1 -2.77 1.23 -0.868
#> 2 -0.969 -1.02 -0.787
#> 3 -2.36 2.54 -1.95
#> 4 -3.68 -0.00565 0.962
#> 5 -4.03 2.07 -0.829
#> 6 -2.59 -0.108 1.32
#> 7 -1.55 -0.651 -0.233
#> 8 -1.18 -0.446 0.134
#> 9 -2.00 -0.485 0.593
#> 10 -0.0356 -0.924 -0.692
#> # ℹ 2,990 more rowssee ?tof_reduce_pca, ?tof_reduce_umap, and
?tof_reduce_tsne for additional details.
Visualization using tof_plot_cells_embedding()
Regardless of the method used, reduced-dimension feature embeddings
can be visualized using ggplot2 (or any graphics
package). tidytof also provides some helper functions for
easily generating dimensionality reduction plots from a
tof_tbl or tibble with columns representing embedding
dimensions:
# plot the tsne embeddings using color to distinguish between clusters
phenograph_tsne |>
tof_plot_cells_embedding(
embedding_cols = contains(".tsne"),
color_col = phenograph_cluster
)
# plot the tsne embeddings using color to represent CD11b expression
phenograph_tsne |>
tof_plot_cells_embedding(
embedding_cols = contains(".tsne"),
color_col = cd11b
) +
ggplot2::scale_fill_viridis_c()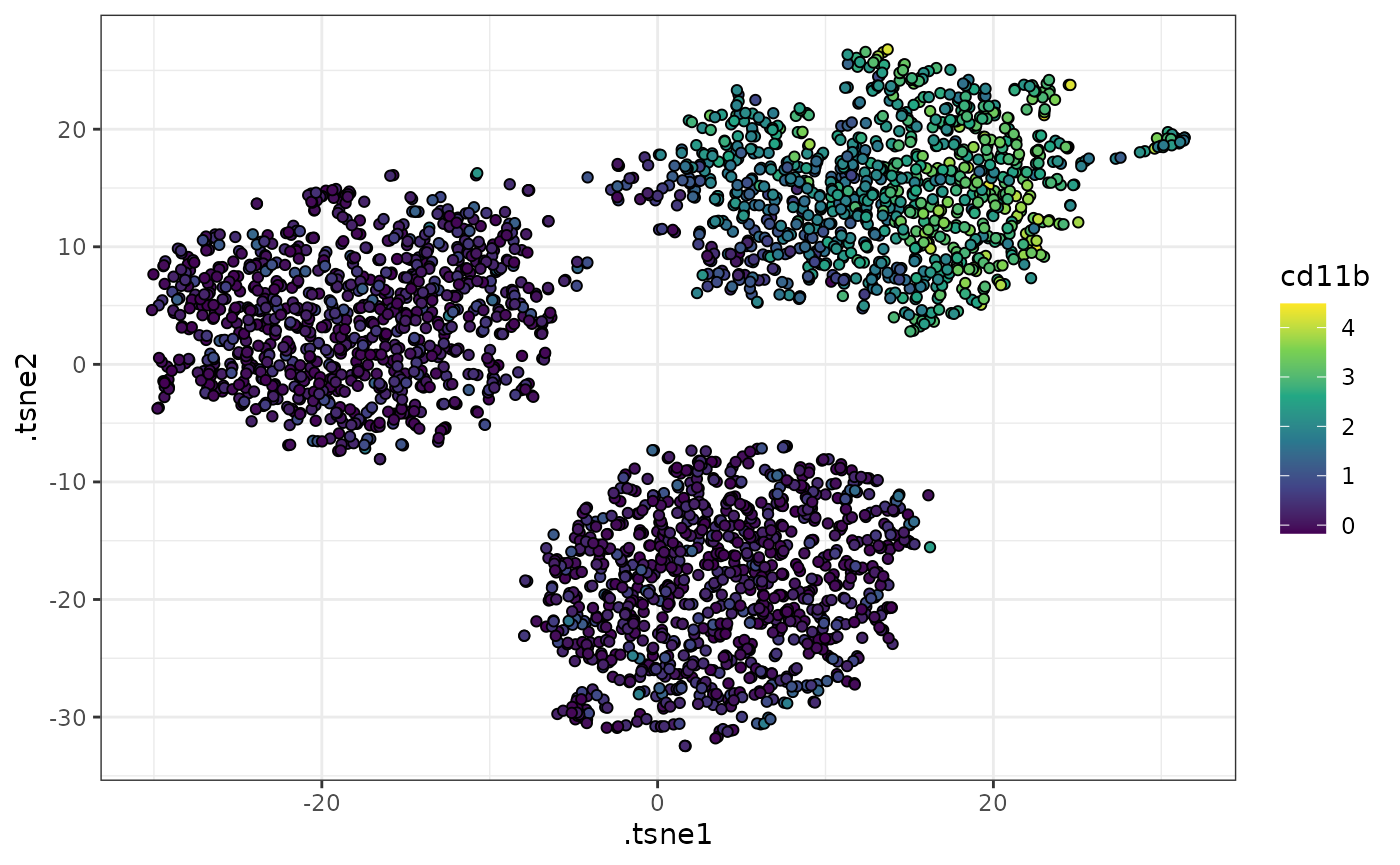
Such visualizations can be helpful in qualitatively describing the phenotypic differences between the clusters in a dataset. For example, in the example above, we can see that one of the clusters has high CD11b expression, whereas the others have lower CD11b expression.
Session info
sessionInfo()
#> R version 4.4.1 (2024-06-14)
#> Platform: x86_64-pc-linux-gnu
#> Running under: Ubuntu 22.04.4 LTS
#>
#> Matrix products: default
#> BLAS: /usr/lib/x86_64-linux-gnu/openblas-pthread/libblas.so.3
#> LAPACK: /usr/lib/x86_64-linux-gnu/openblas-pthread/libopenblasp-r0.3.20.so; LAPACK version 3.10.0
#>
#> locale:
#> [1] LC_CTYPE=C.UTF-8 LC_NUMERIC=C LC_TIME=C.UTF-8
#> [4] LC_COLLATE=C.UTF-8 LC_MONETARY=C.UTF-8 LC_MESSAGES=C.UTF-8
#> [7] LC_PAPER=C.UTF-8 LC_NAME=C LC_ADDRESS=C
#> [10] LC_TELEPHONE=C LC_MEASUREMENT=C.UTF-8 LC_IDENTIFICATION=C
#>
#> time zone: UTC
#> tzcode source: system (glibc)
#>
#> attached base packages:
#> [1] stats graphics grDevices utils datasets methods base
#>
#> other attached packages:
#> [1] ggplot2_3.5.1 dplyr_1.1.4 tidytof_0.99.8
#>
#> loaded via a namespace (and not attached):
#> [1] gridExtra_2.3 rlang_1.1.4 magrittr_2.0.3
#> [4] RcppAnnoy_0.0.22 matrixStats_1.3.0 compiler_4.4.1
#> [7] systemfonts_1.1.0 vctrs_0.6.5 stringr_1.5.1
#> [10] pkgconfig_2.0.3 shape_1.4.6.1 fastmap_1.2.0
#> [13] labeling_0.4.3 ggraph_2.2.1 utf8_1.2.4
#> [16] rmarkdown_2.28 prodlim_2024.06.25 tzdb_0.4.0
#> [19] ragg_1.3.2 purrr_1.0.2 xfun_0.47
#> [22] glmnet_4.1-8 cachem_1.1.0 jsonlite_1.8.8
#> [25] recipes_1.1.0 highr_0.11 tweenr_2.0.3
#> [28] irlba_2.3.5.1 parallel_4.4.1 R6_2.5.1
#> [31] bslib_0.8.0 stringi_1.8.4 parallelly_1.38.0
#> [34] rpart_4.1.23 lubridate_1.9.3 jquerylib_0.1.4
#> [37] Rcpp_1.0.13 iterators_1.0.14 knitr_1.48
#> [40] future.apply_1.11.2 readr_2.1.5 flowCore_2.16.0
#> [43] Matrix_1.7-0 splines_4.4.1 nnet_7.3-19
#> [46] igraph_2.0.3 timechange_0.3.0 tidyselect_1.2.1
#> [49] yaml_2.3.10 viridis_0.6.5 timeDate_4032.109
#> [52] doParallel_1.0.17 codetools_0.2-20 listenv_0.9.1
#> [55] lattice_0.22-6 tibble_3.2.1 Biobase_2.64.0
#> [58] withr_3.0.1 evaluate_0.24.0 Rtsne_0.17
#> [61] future_1.34.0 desc_1.4.3 survival_3.6-4
#> [64] polyclip_1.10-7 embed_1.1.4 pillar_1.9.0
#> [67] foreach_1.5.2 stats4_4.4.1 generics_0.1.3
#> [70] RcppHNSW_0.6.0 S4Vectors_0.42.1 hms_1.1.3
#> [73] munsell_0.5.1 scales_1.3.0 globals_0.16.3
#> [76] class_7.3-22 glue_1.7.0 tools_4.4.1
#> [79] data.table_1.15.4 gower_1.0.1 fs_1.6.4
#> [82] graphlayouts_1.1.1 tidygraph_1.3.1 grid_4.4.1
#> [85] yardstick_1.3.1 tidyr_1.3.1 RProtoBufLib_2.16.0
#> [88] ipred_0.9-15 colorspace_2.1-1 ggforce_0.4.2
#> [91] cli_3.6.3 textshaping_0.4.0 fansi_1.0.6
#> [94] cytolib_2.16.0 viridisLite_0.4.2 lava_1.8.0
#> [97] uwot_0.2.2 gtable_0.3.5 sass_0.4.9
#> [100] digest_0.6.37 BiocGenerics_0.50.0 ggrepel_0.9.5
#> [103] htmlwidgets_1.6.4 farver_2.1.2 memoise_2.0.1
#> [106] htmltools_0.5.8.1 pkgdown_2.1.0 lifecycle_1.0.4
#> [109] hardhat_1.4.0 MASS_7.3-60.2